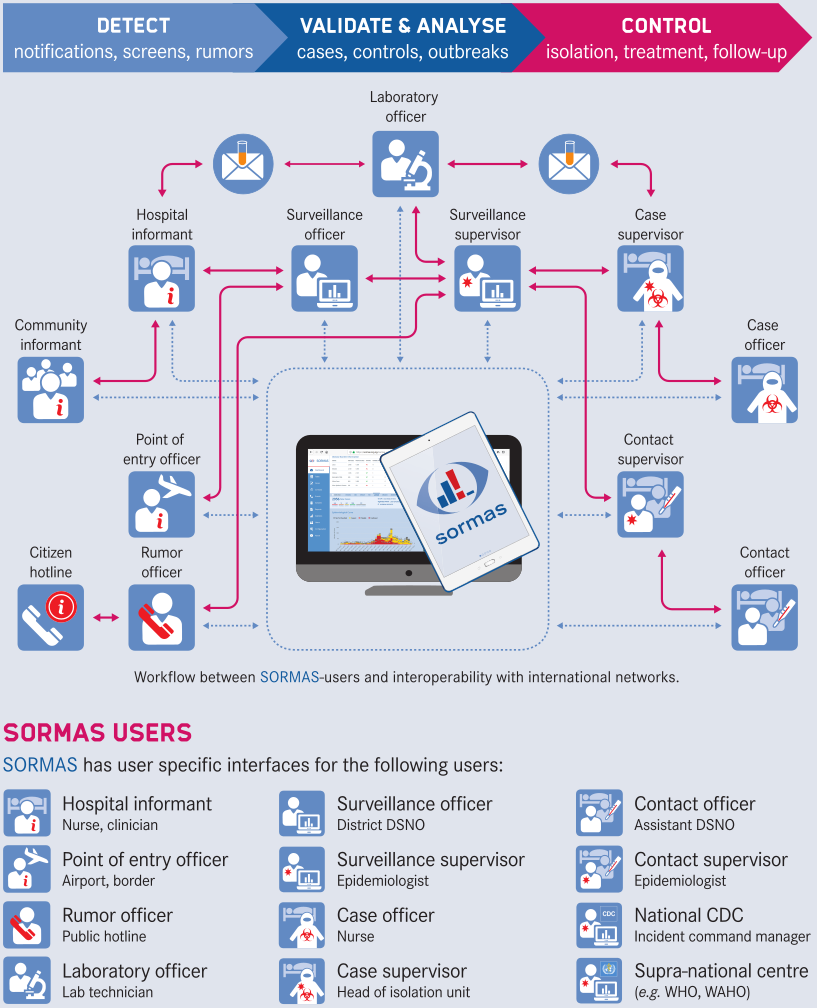
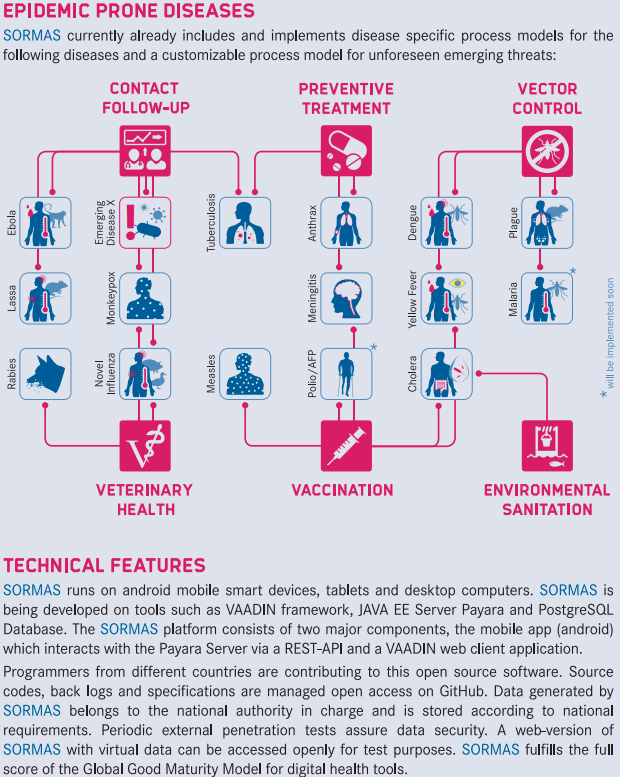
* Zoom in the network diagram to see the contact categories and IDs for person and event nodes
* High risk means Type of contact is Face_to_face_long or Medical_unsave or Touched_fluid or Touched_cloth or Physical_contact else Low risk
* Right click on the network diagram or legend to download or copy.
* The filters are based on complete data. Entities with missing values for the variable used for filtering are not considered
* The data used to generate indicators and figures in this app were obtained from
SORMAS
.